

Optimize the DNA denaturation time and temperature.Mix the reagent stocks and prepared reactions thoroughly to eliminate density gradients that may have formed during storage and setup.Optimize the ratio of the modified nucleotide to dNTP to increase PCR efficiency.Ensure that the selected DNA polymerases are able to incorporate the modified nucleotides.Consider using an additive or co-solvent specifically formulated for a given DNA polymerase (e.g., GC Enhancer supplied with Invitrogen Platinum DNA polymerases).ĭUTP or modified nucleotides in reaction mix.Increase the amount of DNA polymerase, or use DNA polymerases with high processivity.Adjust the annealing temperatures, as high concentrations of PCR additives or co-solvents weaken primer binding to the target.Use the lowest possible concentration when appropriate. Review the recommended concentrations of PCR additives or co-solvents.For example, Pfu DNA polymerase works better with MgSO 4 than with MgCl 2. Check the DNA polymerase’s preference for magnesium salt solutions.The presence of EDTA, other metal chelators, or atypically high concentrations of dNTPs may require a higher Mg 2+ concentration. Optimize Mg 2+ concentration for maximum PCR yields.Increase the amount of DNA polymerase if the reaction mixture contains a high concentration of an additive (e.g., DMSO, formamide) or inhibitors from the sample sources.Review recommendations on the amount of DNA polymerase to use in PCR, and optimize as necessary.Alternatively, set up PCR on ice, or add DNA polymerase last to the reaction mixture.Hot-start DNA polymerases also increase yields of the desired PCR products by eliminating nonspecific amplification. Use hot-start DNA polymerases to prevent degradation of primers by the 3’→5’ exonuclease activity of proofreading DNA polymerases.For long PCR and PCR with degenerate primers, start with a minimum concentration of 0.5 μM.Optimize primer concentrations (usually in the range of 0.1–1 μM).Reconstitute fresh primer aliquots, or obtain new primers if necessary.Aliquot primers after resuspension and store properly.
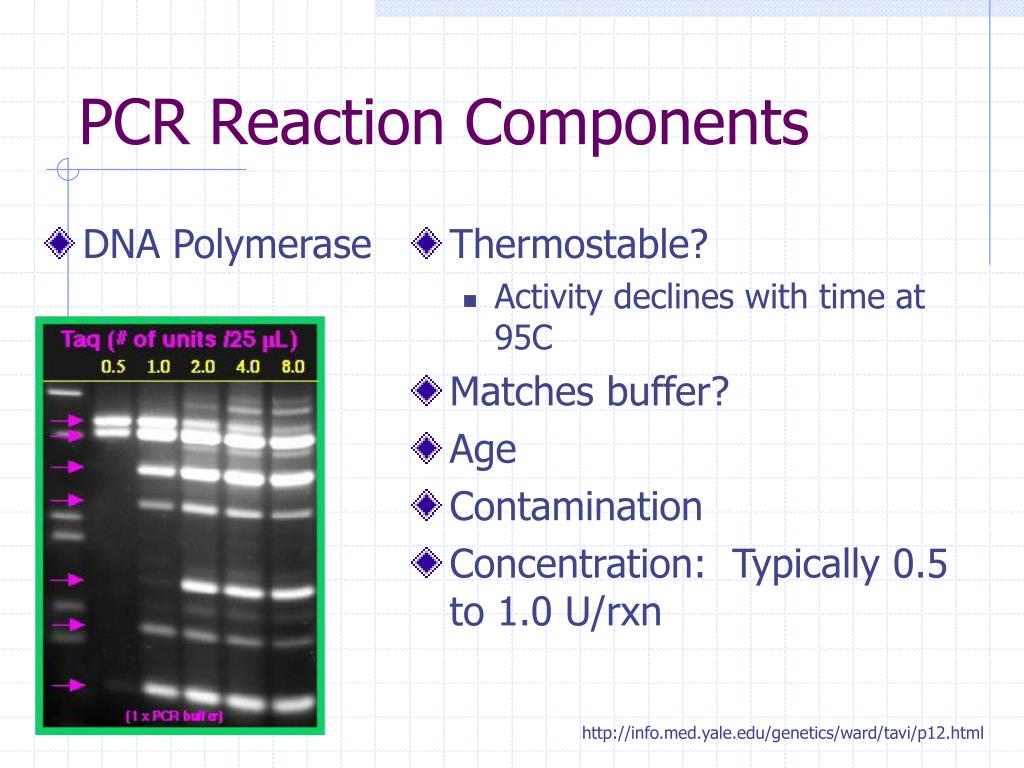
Choose DNA polymerases with high sensitivity for amplification.Examine the quantity of input DNA and increase the amount if necessary.Choose DNA polymerases with high processivity, which display high tolerance to common PCR inhibitors carried over from soil, blood, plant tissues, etc.Re-purify, or precipitate and wash DNA with 70% ethanol, to remove residual salts or ions (e.g., K +, Na +, etc.) that may inhibit DNA polymerases.Ensure that no residual PCR inhibitors such as phenol, EDTA, and proteinase K are present if following chemical or enzymatic DNA purification protocols.
#Pcr troubleshooting band in negative control manual
Consult the user manual and troubleshooting guides to mitigate poor DNA quality. Follow manufacturer recommendations stringently when using purification kits to isolate template DNA.Store DNA in molecular-grade water or TE buffer (pH 8.0) to prevent degradation by nucleases.Evaluate template DNA integrity by gel electrophoresis, if necessary. Minimize shearing and nicking of DNA during isolation.


 0 kommentar(er)
0 kommentar(er)
